Projects
OVERVIEW
We are an evolutionary biology and quantitative genetics lab, and our main study systems are the model system C. elegans and related Caenorhabditis species.
EVOLUTION OF COMPLEX TRAITS
Quantitative traits are determined by many interacting genes, but how do they interact? How does the expression of one allele depend upon another, and how does the expression of an allele depend on the environment? Theoretical work has predicted that perturbations that reveal gene-gene dependencies will also reveal gene-environment dependencies, but empirical data are scarce. We explore these issues to reveal fundamental aspects of complex trait architecture as well as infer how complex traits evolve.
CRYPTIC GENETIC VARIATION IN EMBRYOGENESIS
C. elegans embryos develop with stereotyped precision, even those from genetically distinct strains. However, embryonic genes harbor genetic variation. Do these variants affect how embryogenesis proceeds? To test this question, we knocked down critical embryonic genes, one by one, in many different wild-type strains. We observed substantial variation in embryonic lethality across strains, indicating pervasive cryptic genetic variation in the gene networks for embryogenesis. Such variation has the potential to facilitate adaptive evolution in natural populations; studying it can also reveal the genetic architecture of complex traits.
ENVIRONMENTAL VARIATION AND LOCAL ADAPTATION
By examining patterns of genotypic and phenotypic variation across environmental gradients, we can identify targets of selection and begin to uncover the genetic mechanisms that allow populations to adapt to local conditions. Nematode worms are everywhere and easy to collect; our lab has begun field collection expeditions to the nearby Appalachian foothills in order to characterize the regional distribution of habitats and species. As a genus Caenorhabditis is quite undersampled, a fact that underscores its potential for testing hypotheses in evolutionary genetics.
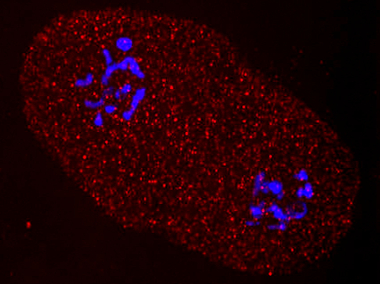
NATURAL VARIATION IN RNA INTERFERENCE
RNA interference (RNAi) is an innate mechanism that induces post-transcriptional gene silencing. The molecular machinery that mediates RNAi is conserved across eukaryotes, and its existence provides experimentalists with powerful reverse genetics tools. However, we and others have discovered that wild C. elegans strains vary in their responsiveness to germline RNAi. Of 55 wild isolates tested, 42 showed incomplete sensitivity to RNAi in the germline, evidenced by increased embryonic survival following RNAi against critical embryonic genes. The genetic and molecular mechanisms that mediate this variation remain unknown, but their discovery may have important consequences for both evolutionary theory and our understanding of epigenetic processes.
EVOLUTION OF DEVELOPMENT
Developmental processes change over time, but do so while also preserving the integrity of the organism. How can such changes take place? Which changes are tolerated, when, and why? In many cases, the cellular and developmental aspects remain relatively stable while the genetic mechanisms undergo re-arrangement, a phenomenon known as developmental system drift. By dissecting the genetic mechanisms and their variation in natural populations of the model system C. elegans, we aim to elucidate the ways in which development changes over long evolutionary timescales.
